1.Click “Species”.
This is a brief introduction to Carthamus Tinctoris L and Arabidopsis thaliana.
(1)Carthamus tinctoris L
(2)Arabidopsis thaliana

2. Click “Search” and go to the page.
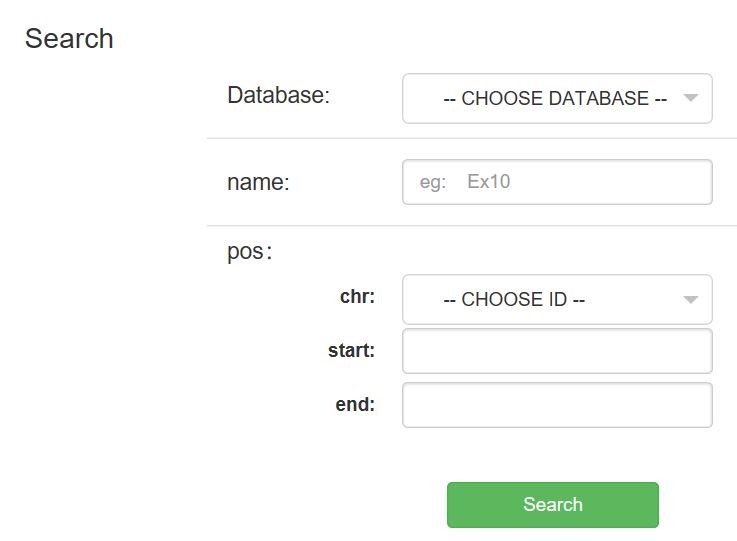
In this “Search” page, the Search Method includes three types: ID, position and keyword. This Genome Database includes Carthamus Tinctoris L and Arabiopsis thaliana. You can find related items, such as Gene, RNA, protein, UTR, DNA, CDs, Gene annotation, GO annotation.
Enter this page,you can search for genes and transcripts based on species, data sets, genome locations, names or keywords. For keywords, you can enter any homologous protein name, KEGG term or EC number, GO term or InterPro term.Under the Search Method option, there are three search methods: ID, location positon or keyword.

(1)Select ID search method under Search Method option, select Arabidopsis thaliana or Carthamus Tinctoris L under Genome Database option, enter the corresponding ID such as AT1G77510, select the items to be searched under Available Tracks option, with Gene, RNA, protein, UTR, DNA, CDs, Gene annotation, GO annotation, and finally click Search to see the results.
Gene results show ID, start location, stop location and gene length, and you can download the result file by clicking download.
1) UTR results show ID, gene location, start location, termination location and length, and can download the result file by clicking download.
2) Protein results show ID, start location, end location and length, and you can download the result file by clicking download.
3) The results of the cDNA display the ID, the starting position, the terminating position, and the basic description type. The result file can be downloaded by clicking download.
4) The results of the mRNA display the ID, the starting position, the terminating position, and the download can be clicked to download the result file.
5) Gene annotation results display and basic description, and you can download the result file by clicking download.
6) GO annotation results show ID, relationship type, GO terminology, GO ID, etc. You can click download to download the result file
The results show like this::

(2) Select the location positon search method under the Search Method option, Arabidopsis thaliana or Carthamus Tinctoris L under the Genome Database option, and enter the corresponding chrom ID, start location and end location, such as chrom ID: superscaffold 1_23_1067230, start: 333, end: 3333.
Under the Available Tracks option, select the items you need to find, including Gene, RNA, protein, UTR, cDNA, CDs, Gene annotation, GO annotation, and click search search to see the search results.
1) Gene results show the corresponding location, starting location, termination location and gene length, and can download the result file by clicking download.
2) UTR results show corresponding locations, gene locations, starting locations, termination locations and lengths, and you can download the results file by clicking download.
3) The CDs results show the corresponding location, starting location, termination location and length, as well as the basic description type, and can click download to download the result file.
4) Protein results show the corresponding location, starting location, termination location and length, and can click download to download the result file.
5) The results of the cDNA display the corresponding position, starting position, terminating position, and basic description type. The result file can be downloaded by clicking download.
6) The MR results show the corresponding position, starting position and terminating position, and can download the result file by clicking download.
7) Gene annotation results show the corresponding location and basic description, and you can download the result file by clicking download.
8) GO annotation results show the corresponding location, relationship type, GO terminology, GO ID, etc., you can click download to download the result file.
The results show like this::

(3) Select the location keyword search method under the Search Method option, select Arabidopsis thaliana or Carthamus Tinctoris L under the Genome Database option, and enter the corresponding keyword, such as Arabidopsis thaliana, keyword: MLP-like. Under the Available Tracks option, select the items you need to find, including Gene, RNA, protein, UTR, cDNA, CDs, Gene annotation and GO annotation. Click Search and see the search results.
1) Gene results show the corresponding location, starting location, termination location and gene length, and can download the result file by clicking download.
2) UTR results show corresponding locations, gene locations, starting locations, termination locations and lengths, and you can download the results file by clicking download.
3) The CDs results show the corresponding location, starting location, termination location and length, as well as the basic description type, and can click download to download the result file.
4) Protein results show the corresponding location, starting location, termination location and length, and can click download to download the result file.
5) The results of the cDNA display the corresponding position, starting position, terminating position, and basic description type. The result file can be downloaded by clicking download.
6) The mRNA results show the corresponding position, starting position and terminating position, and can download the result file by clicking download.
7) Gene annotation results show the corresponding location and basic description, and you can download the result file by clicking download.
8) GO annotation results show the corresponding location, relationship type, GO terminology, GO ID, etc., you can click download to download the result file.
The results show like this::

3.Click on “Population” to go to the page.
You can choose SNP, SSR, QTL, Germplasm.
The genetic map data is merged by …;contains 25,450 markers. The markers include … SSR,6K,60K…
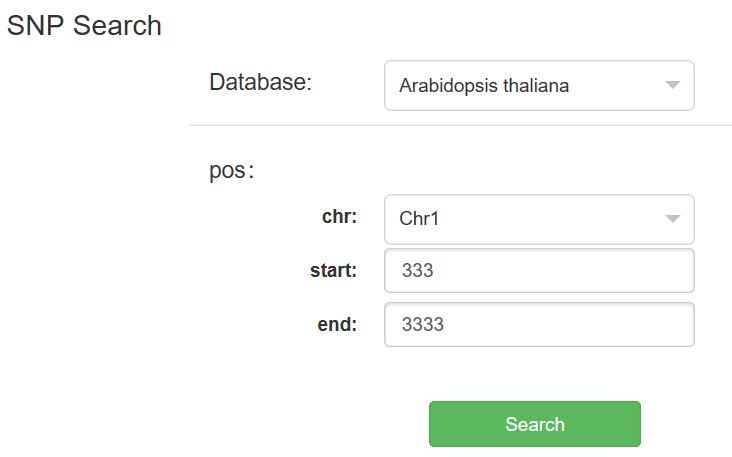
(1)Click SNP, select Arabidopsis thaliana or Carthamus Tinctoris L under the Database option, enter the corresponding chr, start and end, such as chr: superscaffold 8_16_97158024, start: 333, end: 3333, and finally click search search to see the search results.
⑵SSR (Simple Sequence Repeats) marker is a kind of molecular marker technique based on specific primer PCR, which is widely distributed in eukaryotic genome and belongs to the second generation...
Click SSR, select Arabidopsis thaliana or Carthamus Tinctoris L under the Database option, and enter Forward Sequence, Reverse Sequence, Chromosome, SSR markers locus, Coverage, Contig, and SSR markers locus, Coverage, click the corresponding Search button.
Sequence, Chromosome and Contig are shown in Search Results, and scaffoldLong, length, scaffoldShort, mapId, markerpos, leftSequence, rightSequence are shown in each result.

⑶Click on the QTL, select Arabidopsis thaliana or Carthamus Tinctoris L under the Database option, enter name, chr, start and end, and click the corresponding Search button. For example, select Carthamus Tinctoris L, chr: superscaffold 8_16_97158024, start: 333, end: 3333, and click on the corresponding search to see the search results.
⑷Click Germplasm, there are Arabidopsis thaliana, Carthamus Tinctoris L and legume, under Carthamus Tinctoris L, there are many safflower germplasm names, Click to see. For example, click WGHH2063 and you can see it.
4.Click “Tool” to enter the page.
You can choose JBrowse or BLAST.
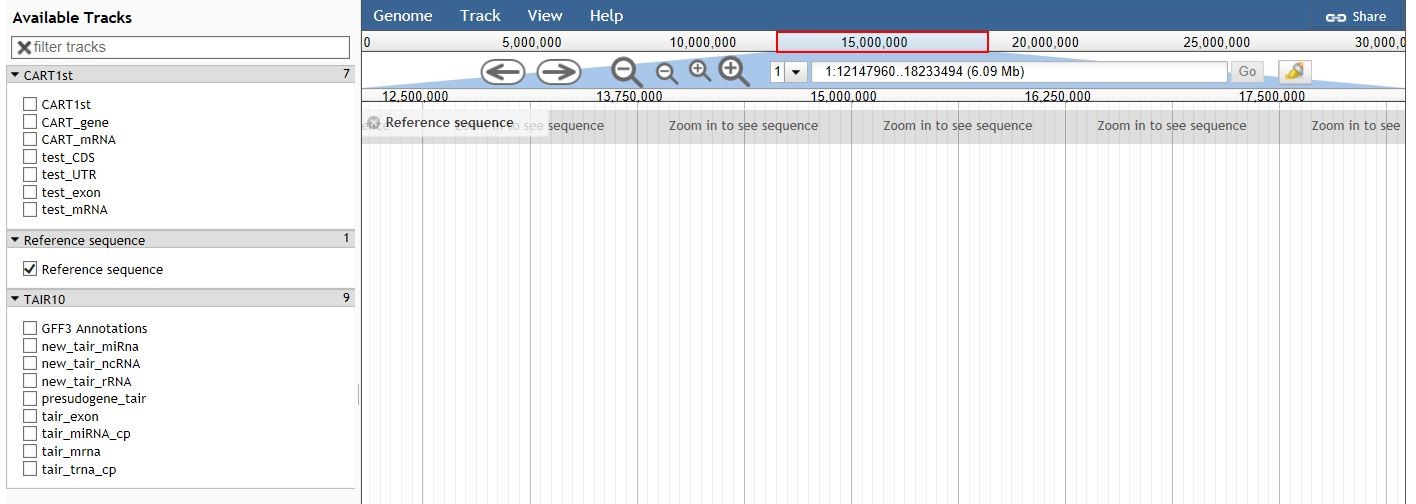
1) Click Jbrowse, click Genome, Open sequence file, open the file, and select CART-gene, CART-RNA, -mrna, cart-mrna, gene, RNA, new-tair-microRNA, new-tair-rRNA, presudene-tair, tair-exon, tair-microNA-cp, tair-mrna, tair¬-trna-cp, CDS, UTR, test-exon, and test-mrna to display the location marker on the map.
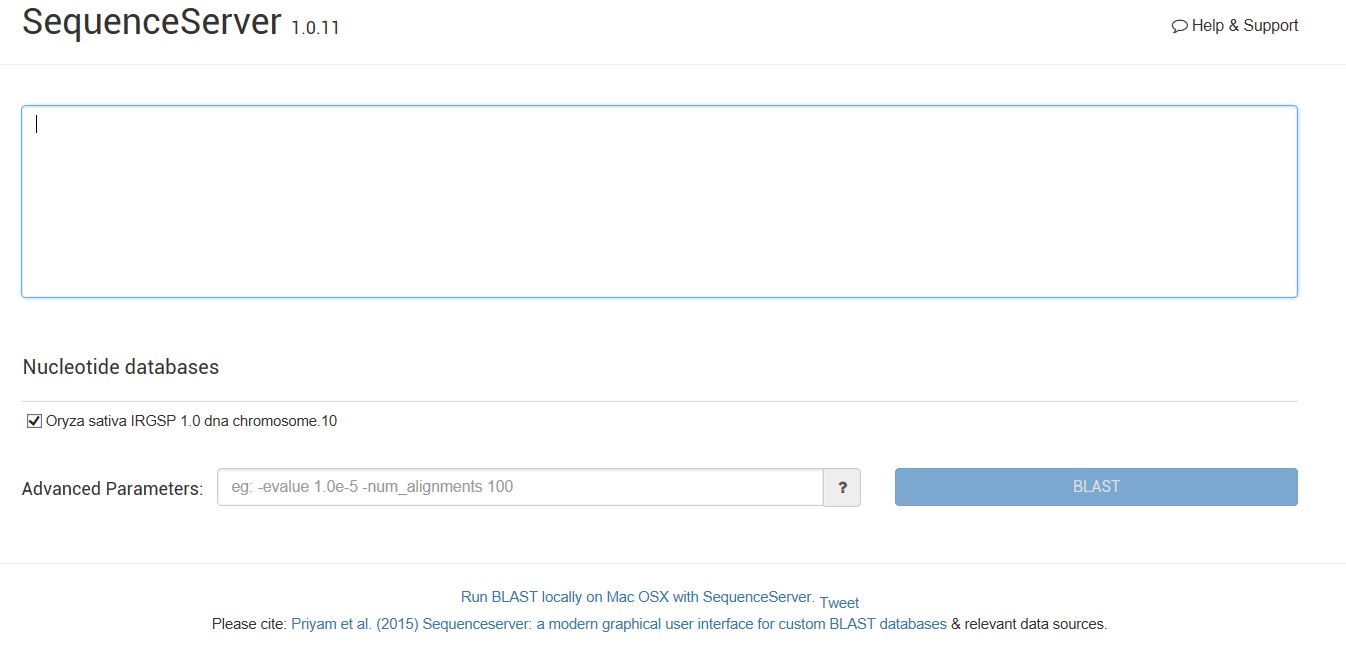
2) Click BLAST, enter the sequence server, paste the query sequence or drag the FASTA format query sequence file, paste query sequence or drag file containing query sequence in FASTA format here.Select Oryza Sativa IRGSP 1.0 DNA chromosome.10 under Nucleotide databases, enter information in Advanced Parameters, eg:- evalue 1.0e-5-num_alignments 100.Finally click the BLAST button.
5.Click “Download” to enter the page.
On the “Download” page, you can download a variety of related data. Other data can be downloaded through the search interface. Only the main data is available through this download page.